Generalized Dirichlet-Multinomial regression and sparse regression
A demo of gen. Dirichlet-Multinomial regression and sparse regression
Contents
Generate generalized Dirichlet-Multinomial random vectors from covariates
clear; % reset random seed s = RandStream('mt19937ar','Seed',1); RandStream.setGlobalStream(s); % sample size n = 500; % # covariates p = 15; % # bins d = 5; % design matrix X = randn(n,p); % true regression coefficients A = zeros(p,d-1); B = zeros(p,d-1); nzidx = [1 3 5]; A(nzidx,:) = 0.5.*ones(length(nzidx),d-1); B(nzidx,:) = 0.5.*ones(length(nzidx),d-1); alpha = exp(X*A); beta = exp(X*B); batchsize = 25+unidrnd(25,n,1); Y = gendirmnrnd(batchsize,alpha, beta);
Fit generalized Dirichlet-Multinomial regression
tic; [Bhat1,Bhat2,stats_gdm] = gendirmnreg(X,Y); toc; display(Bhat1); display(Bhat2); display(stats_gdm); display(stats_gdm.se); display(stats_gdm.wald_pvalue);
Elapsed time is 0.279083 seconds.
Bhat1 =
0.4044 0.4217 0.4496 0.3368
-0.0299 -0.0123 0.0575 -0.0418
0.3943 0.5709 0.5170 0.5504
-0.1911 0.0241 0.0455 0.0265
0.5738 0.6706 0.6396 0.4776
-0.0031 -0.0937 0.0118 -0.0098
-0.0145 0.0940 -0.0978 0.1049
0.0084 -0.1723 0.0355 -0.0211
0.0555 -0.1403 -0.0538 -0.0038
-0.0237 -0.0899 0.0175 0.0732
0.0632 0.1570 0.0418 0.1159
-0.0457 -0.0442 0.0503 0.1275
-0.0062 -0.0604 0.0016 0.0843
0.0931 0.0649 0.0346 0.1222
-0.0544 -0.1110 0.1017 0.0154
Bhat2 =
0.3881 0.3333 0.4765 0.3418
0.0041 -0.0254 0.1981 -0.1771
0.4263 0.5280 0.4249 0.5621
-0.2371 0.0633 0.1171 0.1089
0.5609 0.6077 0.6085 0.5299
-0.0288 -0.0711 -0.0084 -0.0862
0.0126 0.0797 -0.0800 0.1977
-0.0098 -0.2746 0.0930 0.0466
0.0854 -0.1082 -0.1854 -0.0037
-0.0059 -0.0076 0.0470 0.0271
0.1780 0.0620 -0.0126 0.1101
-0.0260 -0.0153 -0.0137 0.0446
-0.0083 -0.0333 0.0510 0.0935
0.0969 0.0572 0.0486 0.0835
-0.0758 -0.0748 0.2051 0.0225
stats_gdm =
struct with fields:
gradient: [15×8 double]
se: [15×8 double]
wald_stat: [1×15 double]
wald_pvalue: [1×15 double]
H: [120×120 double]
observed_information: [120×120 double]
logL: -4.5644e+03
BIC: 9.8746e+03
AIC: 9.3689e+03
dof: 120
iterations: 19
Columns 1 through 7
0.0715 0.0839 0.1085 0.1604 0.0800 0.0908 0.1027
0.0743 0.0837 0.1015 0.1409 0.0787 0.0827 0.1074
0.0667 0.0838 0.1043 0.1407 0.0709 0.0867 0.1074
0.0676 0.0856 0.1065 0.1486 0.0701 0.0833 0.1066
0.0727 0.0925 0.1177 0.1689 0.0772 0.0908 0.1144
0.0678 0.0806 0.1059 0.1400 0.0755 0.0822 0.1011
0.0704 0.0828 0.1042 0.1437 0.0751 0.0802 0.1105
0.0730 0.0878 0.1107 0.1468 0.0790 0.0873 0.1162
0.0714 0.0854 0.1071 0.1453 0.0757 0.0847 0.1101
0.0739 0.0820 0.1113 0.1627 0.0765 0.0824 0.1082
0.0761 0.0914 0.1063 0.1411 0.0808 0.0921 0.1209
0.0722 0.0802 0.1039 0.1423 0.0769 0.0841 0.1074
0.0675 0.0745 0.0975 0.1223 0.0701 0.0820 0.1058
0.0672 0.0794 0.1019 0.1279 0.0779 0.0818 0.1078
0.0745 0.0849 0.1134 0.1527 0.0795 0.0846 0.1141
Column 8
0.1490
0.1400
0.1397
0.1437
0.1654
0.1389
0.1375
0.1461
0.1352
0.1508
0.1418
0.1367
0.1281
0.1232
0.1522
Columns 1 through 7
0.0000 0.0701 0 0.0065 0 0.7036 0.3447
Columns 8 through 14
0.0223 0.0857 0.5563 0.0319 0.5872 0.8417 0.5690
Column 15
0.2119
Fit generalized Dirichlet-Multinomial sparse regression - lasso/group/nuclear penalty
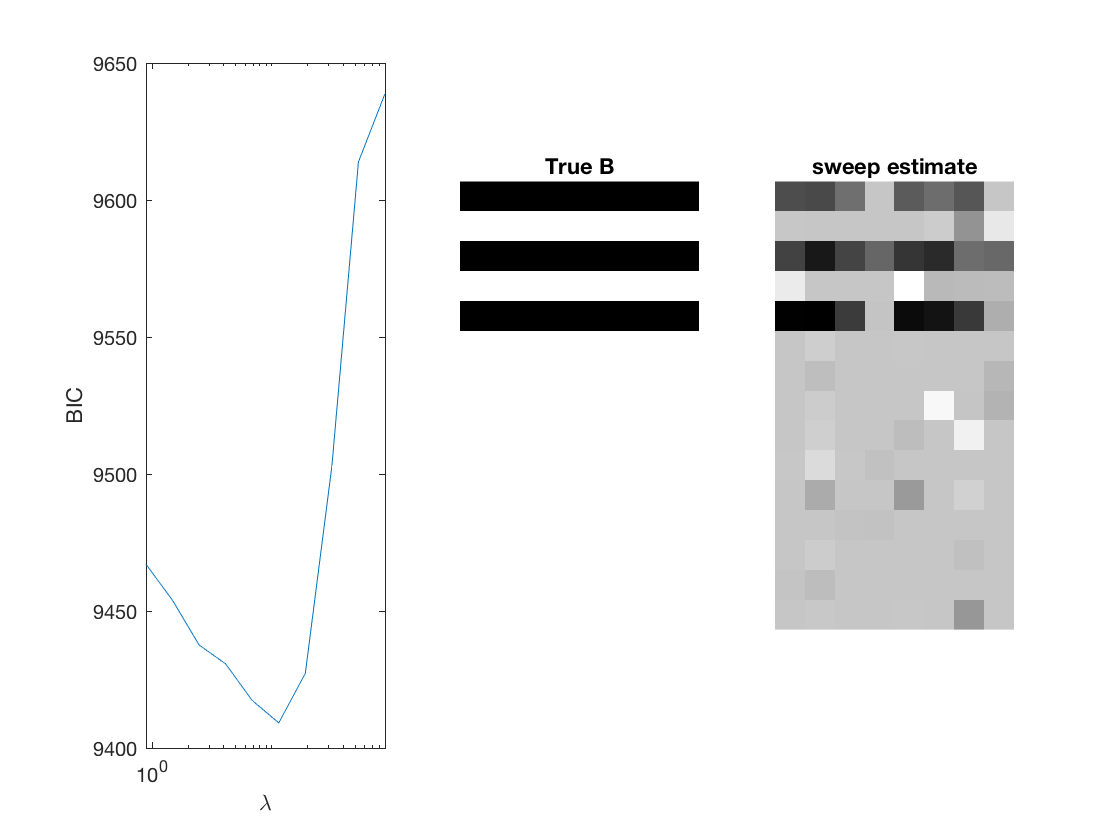
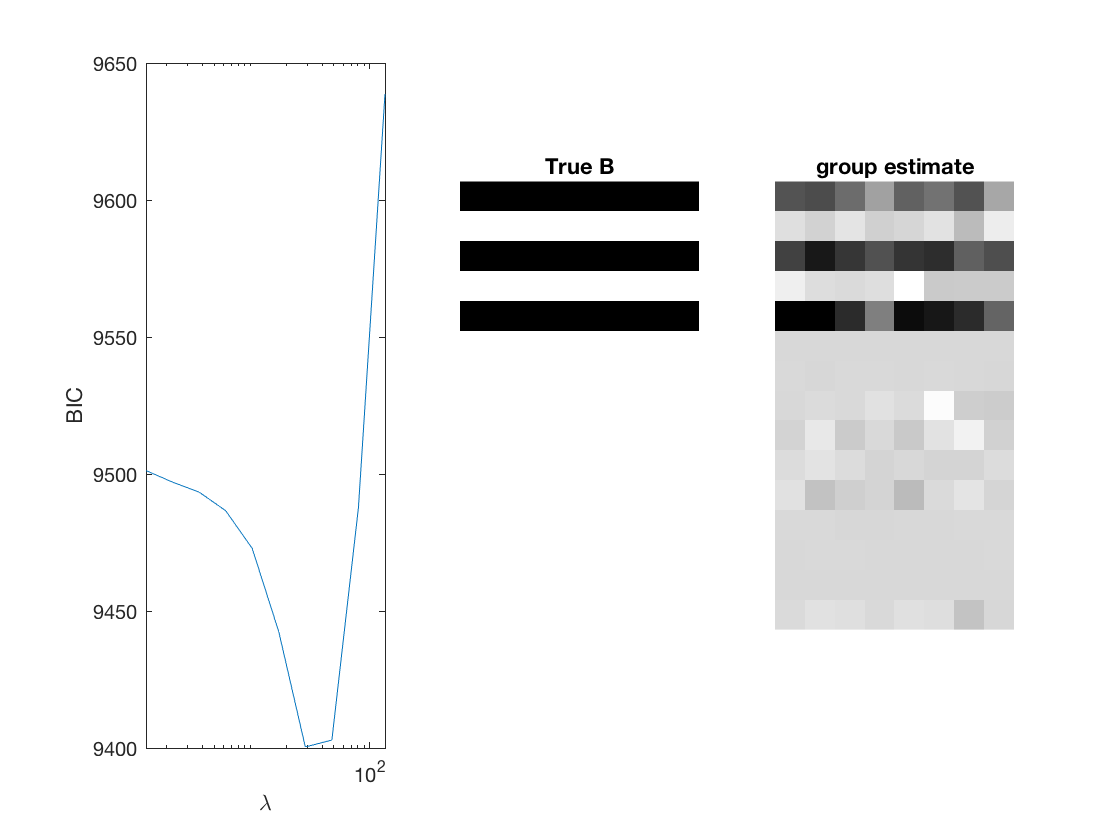
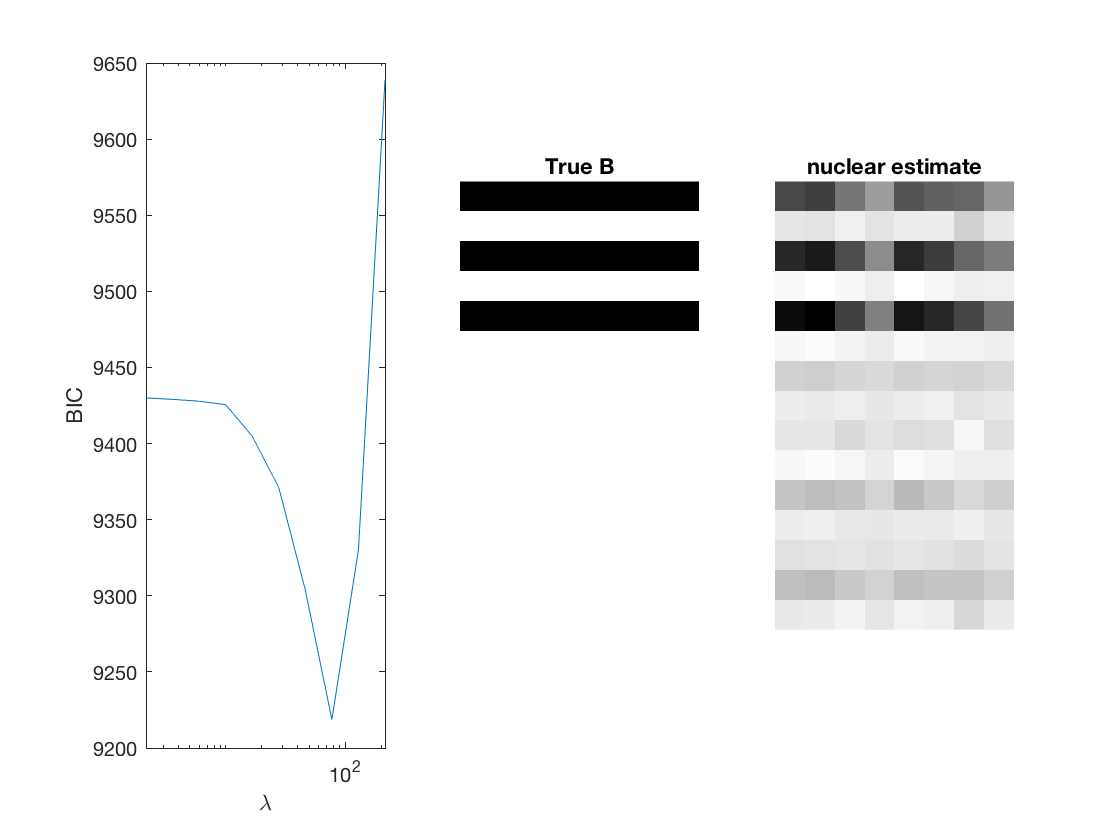
penalty = {'sweep','group','nuclear'};
ngridpt = 10;
dist = 'gendirmn';
for i = 1:length(penalty)
pen = penalty{i};
[~, stats] = mglm_sparsereg(X,Y,inf,'penalty',pen,'dist',dist);
maxlambda = stats.maxlambda;
lambdas = exp(linspace(log(maxlambda),log(maxlambda/100),ngridpt));
BICs = zeros(1,ngridpt);
tic;
for j=1:ngridpt
if j==1
B0 = zeros(p,2*(d-1));
else
B0 = B_hat;
end
[B_hat, stats] = mglm_sparsereg(X,Y,lambdas(j),'penalty',pen, ...
'dist',dist,'B0',B0);
BICs(j) = stats.BIC;
end
toc;
% True signal versus estimated signal
[bestbic,bestidx] = min(BICs);
lambdas(bestidx)
B_best = mglm_sparsereg(X,Y,lambdas(bestidx),'penalty',pen,'dist',dist);
figure;
subplot(1,3,1);
semilogx(lambdas,BICs);
ylabel('BIC');
xlabel('\lambda');
xlim([min(lambdas) max(lambdas)]);
subplot(1,3,2);
imshow(mat2gray(-[A,B])); title('True B');
subplot(1,3,3);
imshow(mat2gray(-B_best)); title([pen ' estimate']);
end
Elapsed time is 0.455057 seconds. ans = 11.4994 Elapsed time is 0.357541 seconds. ans = 29.0023 Elapsed time is 0.439528 seconds. ans = 77.4753